iPSCs and other stem cells
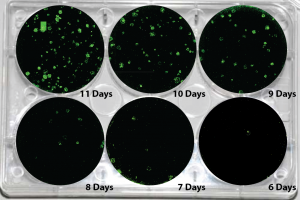
Picture Blog — Human mRNA-Induced Pluripotent Stem Cells Generated in Days
R-iPS Cell FAQ 1:
What phenotypic changes can be observed during a successful reprogramming trial?
About one week out, target fibroblasts should show an involution of fibroblastic processes, and foci or clusters of epitheliod cells—ideally with small nuclei, minimal cytoplasm, and signs of ongoing mitosis—should appear. Colonies with hESC morphology typically start emerging in ~10-14 days.
Making Transfection-Grade mRNA by IVT (In Vitro Transcription)
RNases are an often feared in molecular biology labs because of their high stability and ominous presence in virtually all living systems. Consequently, people who work with RNA are trained to exercise extreme caution to avoid RNA degradation: change gloves often because human hands ooze RNases; use only sterilized labware as microbes may be sources of RNases; for surfaces that can’t be autoclaved, use sprays like “RNase Zap” (SDS- or guanidine-containing solutions). Such cautionary steps are especially necessary when dealing with low abundance RNA samples.
RNAs can be produced by in vitro transcription (IVT), a simple reaction requiring only a DNA template (double-stranded or even single-stranded DNA as long as the promoter region is double-stranded), RNA polymerase (from T7, SP6, or T3 phage), NTPs, and a reaction buffer that provides appropriate salt and pH. Standard NTPs may be replaced with modified ones to either increase stability or to reduce immune-response when transfected into cultured cells. Additionally, a 5’ cap structure may be added during IVT for further stabilizing mRNAs inside the cells post transfection. Using a commercially assembled kit, one can routinely produce 40-50 µg of mRNA from 1 µg of DNA template in a single 20-50 µl reaction.
At such high concentrations, IVT mRNAs are not nearly as sensitive to RNase-mediated degradation as low-abundance samples. The mRNA can be easily observed on agarose gels that are regularly used for DNA, and their integrity can be monitored after transcription or storage. In most cases one distinct band of mRNA from an IVT reaction is obtained as long as a clean DNA template is used. Preparing a good, uniform IVT template is critical to prevent aberrant products. By using high quality templates, IVT mRNA produced in your own lab are often higher in quality than mRNAs purchased from current commercial sources (Figure in Blog shows mRNAs generated by IVT for R-iPSC). Sometimes there are minor bands created during IVT, but they normally do not interfere with the intended uses of the mRNA, and can be purified away with a purification kit (by using a discriminating purification scheme such as Allele Biotech’s Surface Bind RNA Purification, smaller species can be specifically removed, a separate topic for another blog).
Once produced, mRNAs can be stored at -20C for months, or -80C nearly indefinitely.
Development of Cell Lines from iPSCs for Bioassays
The reprogramming of differentiated somatic cells to pluripotency holds great promise for drug discovery and developmental biology. Using immortalized cell lines for drug screening assays has its limitations, such as questionable relevance; and the use of primary cells is often hindered by supply difficulties. Thanks to pioneering work by the Yamanaka, Thompson, and other groups, the feasibility of creating iPSCs has generated an opportunity to provide cell lines with stem cell properties in a virtually unlimited supply [1, 2]. These cells can be derived into different cell types for specific assays, even with patient- or genotype-specific background. Technologies are being developed to produce re-differentiated cells of a number of lineages.
Take cardiomyocytes as an example. There are a number of conventional methods for inducing stem cells into cardiomyocytes: through embryoid body (EB) formation, co-culturing with visceral endoderm-like cell line (END-2), and monolayer caridomyocyte differentiation with defined growth medium and protein factors [3]. A recent publication showed that using appropriate concentrations of BMP4 and activin-A in BSA-containing medium cardiomyocytes might be achieved from iPSCs or ESCs in about 6 days [4].
Transdifferentiation, or direct reprogramming, by introducing a group of 3 cardiomyocyte-specific factors, investigators could directly program cardiac or dermal fibroblasts into cardiomyocyte-like cells [5]. Although much refinement and characterization of these directly reprogrammed cardiomyocyte-like cells, termed iCMs, will be needed before the process can become widely used, this work raised the possibility of quicker and perhaps more efficient ways of generating cells for assays. Similar transdifferentiation has resulted in induced neuron (iN) cells, also by introducing 3 tissue-specific transcription factors [6]. Therefore, it seems that by using defined combinations of tissue-specific transcription factors it is possible to generate cells of different tissue types. It is also possible that by using different, developmental stage-specific transcription activator sets, transdifferentiation can be conducted in a stepwise way and make sure cells at each step is pure. This strategy may be particularly attractive if its efficiency can be improved by the techniques developed for iPSC creation. After all, reprogramming to pluripotency and transdifferentiation to different tissue types must share certain mechanistic steps in their respective processes.
In addition, it has been reported that by briefly overexpressing the Yamanaka iPS factors and controlling growth conditions, mouse fibroblasts could be transdifferentiated up to 40% in 18 days without reversing back to pluripotency [7]. It would be interesting to see if by transient expression of iPS factors via mRNA then switching to cardiomyocyte-specific transcription factors, we can increase the efficiency for direct reprogramming. Use of chromatin-modifying chemicals that were already shown to directly reverse and alter cell fates might also be used to assist direct reprogramming. We believe that a systematic approach for studying these reprogramming aspects should benefit the iPS fields.
1. Takahashi, K. and S. Yamanaka, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 2006. 126(4): p. 663-76.
2. Yu, J., et al., Induced pluripotent stem cell lines derived from human somatic cells. Science, 2007. 318(5858): p. 1917-20.
3. Vidarsson, H., J. Hyllner, and P. Sartipy, Differentiation of human embryonic stem cells to cardiomyocytes for in vitro and in vivo applications. Stem Cell Rev, 2010. 6(1): p. 108-20.
4. Elliott, D.A., et al., NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods, 2011.
5. Ieda, M., et al., Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell, 2010. 142(3): p. 375-86.
6. Pang, Z.P., et al., Induction of human neuronal cells by defined transcription factors. Nature, 2011. 476(7359): p. 220-3.
7. Efe, J.A., et al., Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol, 2011. 13(3): p. 215-22.
New Products of the week: T7 RNA Polymerase, high quality for demanding in vitro transcription requirements.
Promotion of the week: GFP-Trap, buy 2 of any package and get 1 of equal or less value free. Use code FreeTrap, follow deals quickly on Facebook.
Creating ground-state human iPSCs
Murine pluripotent stem cells can exist in two distinct states, blastocyst-derived LIF-dependent embryonic stem cells (ESCs) and epiblast-derived bFGF-dependent stem cells (EpiSCs). Murine ESCs and similar iPSC lines are more of the “ground-state” in terms of developmental status, as reflected by the lack of X chromosome inactivation in female cells and their abilities to pass as single cells. Human iPSCs, like human ES cells, are more similar to mouse EpiSCs. Unfortunately these human pluripotent stem cells are difficult to genetically manipulate, e.g. knockin or knockout. They also grow slowly, with doubling time averaging 36 hours. In order to create ground-state human iPSCs, several approaches have been tested, including reprogramming iPSC-derived fibroblasts, continuously expressing 5 iPS factors (Oct4, Sox2, Nanog, c-Myc, and Klf4), or using chemicals to inhibit chromatin modifying enzyme HDAC. While these approaches succeeded to certain degrees, the resulting cell lines seem to have some limitations, such as limited passage numbers.
Retinoic acid (RA) signaling is involved in many aspects of embryonic development. RA receptor (RAR), together with one of its heterodimerization partners, steroid hormone receptor Lrh-1, was recently found to be able to synergize with the 4 common iPS factors (Oct4, Sox2, Klf4, and c-Myc) to induce mouse and human fibroblasts into ground-state iPSCs. The pluripotent cells created by the so-called F6 factor combination show no X chromosome inactivation if from female origin, can fully activate the endogenous Oct4 promoter, express Rex1 (which is specific to mouse ESCs, not EpiSCs), and grow with a 16 hour doubling time. All these mouse ESC-like features were achieved without detectable expression of the exogenous factors once iPSC colonies formed, indicating transient F6 expression is capable of effectively initiating endogenous stem cell factors. Remarkably, these stem cells can maintain their undifferentiated status in mouse ESC medium for 50 passages or more. This work, published this month in Proceedings of National Academy of Science USA [1], provided the stem cell research and application field with a very desirable choice of human stem cells.
As opposed to ~16 days with F4, it appears that the time required to induce adult fibroblasts into pluripotent stem cells is as short as 4 days if F6 factors are introduced on a murine stem cell virus (MSCV) vector with an integrated piggyback transposon. As the authors noted in their discussion, the speed-up benefit should be particularly advantageous for transient transfection approaches such as mRNA reprogramming. The bottom line from this paper and the engineered factor papers (see the previous AlleleBlog article under “iPS and other Stem Cells”) is that iPSC reprogramming is only going to get faster, which means that hopefully in the near future creating iPSCs will become a routine experiment as easy as a simple transfection.
Wang, W., J. Yang, et al. (2011). “Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1.” Proc Natl Acad Sci U S A.
New Products of the week: ARCA, modified cap analog for in vitro transcription of mRNA.
Promotion of the week: Friday special this week, 15% off all iPS viral particle products if using code “ViraliPS” when ordering online at allelebiotech.com, by email, or fax.
Fusion of the Transcription Domain to iPS Factors Radically Enhances Reprogramming
Induced pluripotent stem cells (iPSCs) can be achieved through introduction of a small group of stem cell specific transcription factors. Ever since this was first demonstrated by Takahashi and Yamanaka, there have been relentless efforts for improving the efficiency of this generally inefficient process. There is also a general opinion that iPSCs are different from each other and from embryonic stem cells (ESCs) in various aspects, depending on the method of the induction. As a result, another focus of the reprogramming field has been to find ways for creating iPSCs that are as close to ESCs as possible. One of the parameters for defining stem cell status is their epigenetic characters; epigenetic changes have been demonstrated to occur during reprogramming of subsequent differentiation.
In fact, it seems that reprogramming can be largely described as a process composed of chromatin remodeling and specific transcription activation. Strong transcription activators are known to effectively recruit multiple chromatin remodeling complexes when exerting their functions. A good example is MyoD, a master transcription factor for skeletal myogenesis that can “single-handedly” switch (transdifferentiate) the fate of differentiated cells. Hirai et al. speculated that since MyoD is such a strong transcription factor, it may be able to increase chromatin accessibility to iPS factors if fused together. When transduced on retroviral vectors, Oct-TAD (Transcription Activation Domain) of MyoD, in combination with Sox2 and Klf4, increased the number of iPSC colonies by 40-fold. Additionally, these iPSCs appeared to quickly adopt stem cell gene expression profiles, days faster than when traditional Oct4, Sox2, c-Myc, and Klf4 were used; and sometimes the levels of pluripotency genes even exceeds those seen in ESCs. Amazingly, when using the fusion assisted method some colonies are formed without the help of feeder cells, a requirement of ESCs grown in similar medium. Does this mean that these iPSCs can even be more “stem-like” than embryonic stem cells?
Like MyoD, VP16, also widely known for its strong transcription activation domain, when fused to iPS factors, was shown to exhibit a similar stimulation effect on reprogramming. Although the details of the fusion arrangements and specificity appear to differ between MyoD and VP16, the fact that two research groups could achieve similar results using comparable strategies provides a good argument that other labs should at least consider this method when creating mouse or human iPSCs. Previously in our blog we have discussed using iPS factor mRNAs, a method originally developed by Warren et al., for substantially shortening the time required for reprogramming and making it more robust across cell types and media conditions. If the new TAD-fusion factors are used also in the mRNA format, then the protocol might be further shortened and simplified. If successful, this non-integrating approach could become a dominant method in the field, even making competitive non-integrating method such as Sendai and plasmid-based miRNA irrelevant.
New product of the week:SurfaceBind RNA purification system, higher capacity and simpler procedure than Qiagen or Ambion’s comparable products, particularly suitable for mRNA cleanup after in vitro transcription.
Promotion of the week: 30% off RNA SurfaceBind Purification kits. To redeem this offer email the code PURIFY to oligo@allelebiotech.com.
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008