iPSCs and other stem cells
Press Release: Allele Biotechnology & Pharmaceuticals Closes Purchase of cGMP Facility for Production of Clinical-Grade Cells for Cell Therapy Applications
SAN DIEGO–(BUSINESS WIRE)–Allele Biotechnology & Pharmaceuticals, Inc. (“Allele”), a leader in the development of specialized cells for pharmaceutical drug discovery and regenerative medicine, today announced that it has closed the purchase of a new facility intended for its cGMP (current good manufacturing practices) production of clinical-grade cells for cell therapy applications.
The 18,000 square-foot facility, located near the main headquarters of Allele in San Diego, California, will be the center of production of human induced pluripotent stem cells (hiPSCs) using Allele’s proprietary synthetic mRNA platform, a technology that generates hiPSCs with neither the random integration of foreign DNA nor the use of whole virus or virus-based elements, drawbacks that are common to other technologies for making hiPSCs. Such “footprint-free” cells will be produced by Allele for industrial and academic partnerships, as well as Allele’s own efforts in the area of cellular therapeutics.
hiPSCs, as cells that have the potential to become any cell in the human body, hold great promise for therapies that can alleviate or cure human disease. Towards this end, Allele has recently made a number of advances regarding the differentiation of hiPSCs towards cells of specific lineages, such as neural progenitor cells, neurons, astrocytes, mesenchymal stem cells, cardiomyocytes, skeletal muscle cells, hepatocytes, and adipocytes, including brown fat cells. These cells would also be produced in the cGMP facility when intended for specific therapies.
“This dedicated facility will help us to realize a number of our visions in bringing the benefits of pluripotent stem cells to society,” said Jiwu Wang, Ph.D., President and CEO of Allele. “The first step in helping people in need with all the stem cell technologies developed in labs is to clear a path to move them from bench to bedside, which requires high-quality, controlled production that can be monitored by the FDA. Together with our licensees, drug development partners, investors, and individuals who would like to participate in banking hiPSCs for research and therapy, we anticipate even faster pace in our business development in this area.”
Contacts
Allele Biotechnology & Pharmaceuticals, Inc.
Jiwu Wang, 858-587-6645
info@allelebiotech.com
Press Release: Ocata Therapeutics Licenses Induced Pluripotent Stem Cell Technology from Allele Biotechnology and Extends Leadership Position in Cell Therapy Capabilities
Ocata Announces Proof-of-Concept Results in Restoring Vision and Prevention of Blindness
MARLBOROUGH, Mass., Mar 24, 2015 (BUSINESS WIRE) — Ocata Therapeutics, Inc. (“Ocata” or “the Company”; NASDAQ: OCAT), a leader in the field of Regenerative Ophthalmology™, today announced that it has entered into a definitive agreement with Allele Biotechnology & Pharmaceuticals, Inc. of San Diego, CA (“Allele”) to access Allele’s proprietary technology for generating human induced pluripotent stem cells (“iPSCs”).
“This agreement with Allele is part of our strategy to broaden our technology platform and increase our leadership in regenerative ophthalmology,” said Paul Wotton Ph.D., President and CEO of Ocata. “Ocata can now take advantage of induced and embryonic pluripotent stem cells to produce commercially viable human tissue for transplantation. We recently confirmed proof of concept in creating photoreceptors capable of preventing blindness and restoring vision in established animal models. Data from these studies will be published later this year.”
Since Dr. Yamanaka discovered how to generate iPSCs in 2007 there has been tremendous enthusiasm about the potential to use these pluripotent cells to develop commercially viable therapies. Despite many efforts to develop iPSC derived therapies in the same scalable and reliable way as embryonic stem cells, many of those efforts have been unsuccessful due to issues relating to the growth capacity, differentiation potentials and epigenetic properties of iPSCs. The “footprint-free” reprogramming technology developed by Allele potentially offers a reliable and scalable process for producing iPSCs with superior properties and is a major step for translation of iPSC technology to practical clinical use. These iPSCs can potentially be used to manufacture millions of treatment doses as off-the-shelf therapies for any patient.
“We have had a strong leadership role in this area,” said Robert Lanza, M.D., Chief Scientific Officer of Ocata. “Ocata has extensive experience and patent rights in generating both ocular and non-ocular cell types from human iPSCs. We have painstakingly and patiently evaluated many different iPSC technologies and selected the Allele technology only after we were satisfied and confident that this represented the best of all approaches and could permit us to generate transplantable tissues that would be potentially safe in human patients. In our hands, the iPSCs we are generating are comparable to our embryonic stem cells in those features required for use in potential human therapies.”
“It is particularly rewarding to us that Ocata, a company whose understanding of the science and regulatory requirements in this space is unparalleled, has selected the iPSC technology developed at Allele for application in its own pipeline,” said Jiwu Wang Ph.D., President and CEO of Allele. “It only serves to confirm our belief that our iPSC platform is a solution to what otherwise have been unresolved issues associated with the maturation of iPSCs to a fully functional state. The ability to predictably derive stable iPSC lines without using any viral element or foreign DNA enables both fundamental scientific research and clinical applications, which has been the mission of Allele Biotechnology from its inception.”
For Contact at Allele:
oligo@allelebiotech.com,
P 858-587-6645, 800-991-RNAi(7624)
F 858-587-6692
For full release with Ocata contact, see MarketWatch
Allele’s SBIR Grant to Develop All-RNA CRISPR
Precise engineering of the genomes of mammalian cells enabled biological and medical applications researchers had dreamed of for decades. Recent developments in the stem cell field have created even more excitement for genetically modifying genomes because it enables delivering more beneficial stem cell-derived therapeutic cells to patients [1]. For instance, by correcting a gene mutation known to be critical to Parkinson’s disease, LRRK2 G2019S, in patient-specific iPSCs (induced pluripotent stem cells), it appeared possible to rescue neurodegenerative phenotypes [2].
Significant amount of fund and energy had been invested in technologies such as ZFN and TALEN, however, judging from the explosion of publications and business activities in just about 2 years since the illustration of its mechanism (just today, Jan 8th, 2015, Novartis announced CRISPR collaborations with Intellia, Caribou, applying it in CAR T cell and HSCs), the CRISPR/cas system is the rising star. This system uses a guide RNA to direct the traffic of a single nuclease towards different targets on a chromosome to alter DNA sequence through cutting. The nuclease, cas9, can be mutated from a double-stranded DNA endonuclease to a single-strand cutter or a non-cutting block, or further fused to various functional domains such as a transcription activation domain. This system can also be used to edit RNA molecules.
A weak spot on the sharp blade of CRISPR is, like any methods for creating loss-of-function effects (RNAi if you remember), the potential of off-target effects. While they can never be completely avoided, with the ever growing popularity of deep sequencing, at least we can know all unintended changes on the edited genome. Almost a perfect storm! As an interesting side story, when we at Allele Biotech first saw the paper in Science describing the CIRPSR/cas system [3], we immediately wrote an SBIR grant application for applying the bacterial system to mammalian cells. The first round of review in December 2012 concluded that it would not work due to eukaryotes’ compact chromatin structures. Of course, the flurry of publication in early 2013, while our application was being resubmitted, proved otherwise. The good news is, Allele Biotech still received an SBIR grant from NIGMS in 2014. Unlike most of the genome editing platforms known in the literature, our goal was to build an all-RNA CRISPR/cas system, thereby with higher potency, less off-target effects, and, as a footprint-free platform, more suitable for therapeutic applications. This system will be combined with our strengths in iPSC and stem cell differentiation, fluorescent protein markers, and deep sequencing based bioinformatics to improve cell therapy and cell based assays.
1 Urnov, F.D., et al., Genome editing with engineered zinc finger nucleases. Nat Rev Genet, 2010. 11(9): p. 636-46.
2 Reinhardt, P., et al., Genetic Correction of a LRRK2 Mutation in Human iPSCs Links Parkinsonian Neurodegeneration to ERK-Dependent Changes in Gene Expression. Cell Stem Cell, 2013. 12(3): p. 354-67.
3 Jinek, M., et al., A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science, 2012.
Combining mRNA-Mediated Phenotype Rescue and CRISPR-Created Isogenic Genome
There has been a good volume of publications on using patient-specific iPSCs for disease modeling. Among them, a recent study by Wang et al. published in Nature Medicine is unique because it not only created cardiomyocytes from both Barth Cardiomyopathy patient and wildtype samples for functional analysis, but also combined some of the most exciting new technologies to strengthen the correlation between gene change and disease.
First, functional rescue by mRNA transgene. After mRNA that encodes wild-type cardiolipin aclation enzyme encoding gene tafazzin (TAZ) was transfected into Barth iPSC-CM cells, their defects in mitochondrial functions were corrected. Second, loss of function by genome editing. When CRISPR was used to make genome changes in wildtype cells that mimicked the disease-specific mutation, we recreated the patient’s iPSC-CM phenotype in otherwise wildtype cells. Third, next generation sequencing to confirm genomic changes. And forth, the cardiomyocyte contractibility was assayed on bioengineered chips.
This paper should set an example of how patient iPSCs should be used to create disease models to the fullest extent of usefulness and reliability. We are true believers of the idea that technology development empowers the advancement of science.
“Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies.” Wang, G., McCain, M.L., Yang, L., He, A., Pasqualini, F.S., Agarwal, A., Yuan, H., Jiang, D., Zhang, D., Zangi, L., Geva, J., Roberts, A.E., Ma, Q., Ding, J., Chen, J., Wang, D.Z., Li, K., Wang, J., Wanders, R.J., Kulik, W., Vaz, F.M., Laflamme, M.A., Murry, C.E., Chien, K.R., Kelley, R.I., Church, G.M., Parker, K.K., Pu, W.T. (2014) Nature Medicine, Jun;20(6):616-23. doi: 10.1038/nm.3545. Epub 2014 May 11.
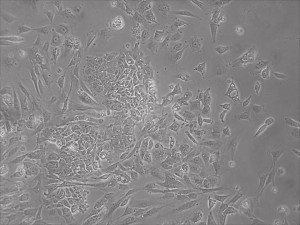
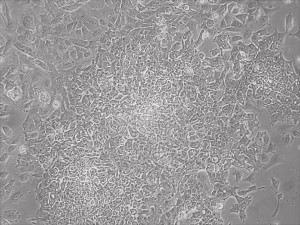
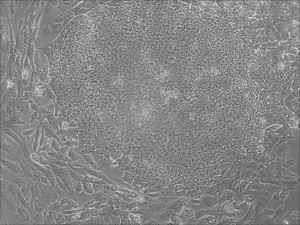
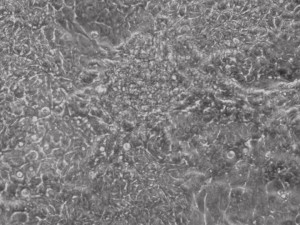
Appearance of iPSCs–Different Reprogramming Stages within the Same Well
Previously scientists at Allele Biotech have reported near uniform conversion of human fibroblasts using our proprietary mRNA mixtures. The first picture below shows a well of cells after 7 days of growing fibroblasts with the new Allele mRNA mix.
This month, by adjusting the mRNA dose while testing Allele’s own reprogramming medium formulation, we observed various stages of cells going through the transition in the same well (see pictures 2 to 5). All stages of reprogramming typically observed over a span of weeks can actually be seen within 1 well of a 6-well plate when we treated human fibroblasts at half the dose of our standard mRNA mix, on day 10, and using Allele Biotech’s new formulation of reprogramming medium.
(1) Warren, Ni, Wang, and Guo 2012 (pdf download)
Previous bulk conversion on Day 7 of reprogramming at full dose mRNA, improved upon our published efficiency (1)
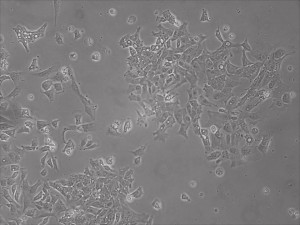
Reprogramming en masse: post mesenchymal-to-epithelial (MET) transition cells start to become iPSCs without surrounding fibroblasts (as opposed to the above figure)
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008