mRNA reprogramming
Appearance of iPSCs–Different Reprogramming Stages within the Same Well
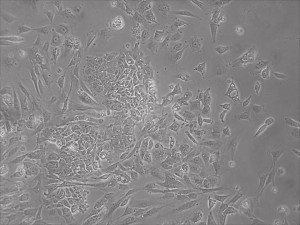
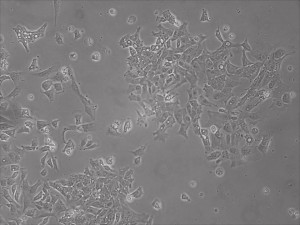
Previously scientists at Allele Biotech have reported near uniform conversion of human fibroblasts using our proprietary mRNA mixtures. The first picture below shows a well of cells after 7 days of growing fibroblasts with the new Allele mRNA mix.
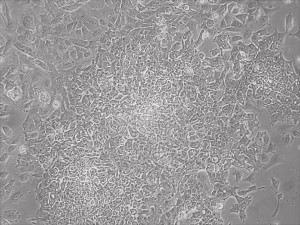
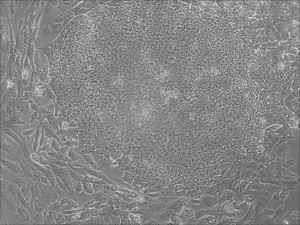
This month, by adjusting the mRNA dose while testing Allele’s own reprogramming medium formulation, we observed various stages of cells going through the transition in the same well (see pictures 2 to 5). All stages of reprogramming typically observed over a span of weeks can actually be seen within 1 well of a 6-well plate when we treated human fibroblasts at half the dose of our standard mRNA mix, on day 10, and using Allele Biotech’s new formulation of reprogramming medium.
(1) Warren, Ni, Wang, and Guo 2012 (pdf download)
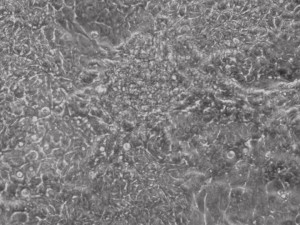
Previous bulk conversion on Day 7 of reprogramming at full dose mRNA, improved upon our published efficiency (1)
Reprogramming en masse: post mesenchymal-to-epithelial (MET) transition cells start to become iPSCs without surrounding fibroblasts (as opposed to the above figure)
Picture Blog: How Do You Like Your iPSCs, Clonal or Bulk-Conversion?
Reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs) is commonly considered a stochastic process, i.e. with randomness, which offers an excuse for the commonly seen low efficiency and low constancy of making iPSCs. We have demonstrated time and again that by using potent mRNA cocktails, the majority of the fibroblasts seeded in a well can be converted into pluripotent stage in a nearly synchronized manner (Warren et al. 2012, Warren and Wang 2013, and this Allele Picture Blog series). mRNA molecules can function robustly yet transiently while avoiding the need of entering the nucleus, a bottle-neck for all DNA-based vehicles.
Other researchers are used to the idea of clonal expansion partly because isolating iPSCs from “clones” was a common step during reprogramming using viruses or other low efficiency methods, even though those clones were not necessarily from single precursor cells. This week, the Allele iPSC team developed a new way of managing our mRNA reprogramming that allowed us to achieve clonal iPSCs that appear to be a lot purer and more likely true clones compared to previous reports, without compromising any of the main benefits of our protocol, e.g. feeder-free, xeno-free, footprint-free, very fast and highly efficient. This work is currently supported by an NIDA/NIH grant to Dr. Jiwu Wang at Allele Biotech.
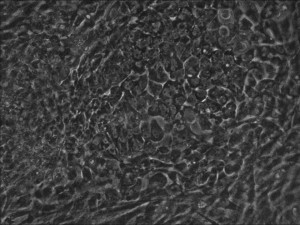
Traditional bulk-conversion by the Allele mRNA reprogramming protocol developed by Warren et al. The picture shows large patches of cells becoming stem cells almost overnight around the 9th day of adding mRNA-cocktail supplement to the media.
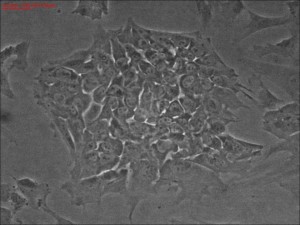
Clonal iPSC formation using a modified mRNA reprogramming protocol. The picture shows a typical clone of stem cells that originated from likely single cells.
Warren, Ni, Wang, and Guo, Scientific Reports, 2012
Warren and Wang, Current Protocols, 2013, in press
Path to Better Drugs through Disease-Specific iPSCs
Induced human pluripotent stem cells
The recent finding that pluripotency, the ability to differentiate into all cell types typically associated with embryonic stem cells, can be induced in somatic cells may be the molecular equivalent of the discovery of antibiotics or vaccines in the last century [1].
iPSC-based disease modeling
Recent studies have described the generation of induced pluripotent stem cells (iPSCs) from patients with a full range of genetically inherited or sporadic diseases, and in vitro differentiation of these iPSCs to cell types relevant to the disorder with certain disease features.
Example 1 (out of ~20): Progressive motor neuron loss during differentiation of iPSCs derived from spinal muscular atrophy (SMA) patients, reflecting developmental loss seen in the disease.
Example 2: iPSCs made from RETT syndrome give rise to glutamatergic neurons with fewer synapses than controls, a better treatment was found from a panel of candidates based on this model.
Example 3: Neurons differentiated from iPSCs that have been derived from early or late onset Alzheimer’s disease were shown to display different properties and potential interference points.
The identification of novel pathways or drugs that could prevent disease is the ultimate goal of the iPSC-based disease modeling approach.
Major steps towards efficient iPSC disease modeling
The first hurdle for feasible application of patient-specific disease modeling is to achieve efficient generation of iPSCs from large cohorts of patients quickly and at a low cost while eliminating “clonal variations”. As described in a recent publication [2], the Allele Biotechnology team has shown that human fibroblasts can be converted to stem cells in just over a week, achieving bulk conversion efficiency without any chromosome modifications. The process is also xeno-free and feeder-free, enabling both fundamental scientific research and clinical applications.
The next major advancements required for disease modeling are robust lineage-specific differentiation protocols that provide a large number of desired cells for drug testing and screening. Cardiomyocytes derived from iPSCs have been the best known example of large expansion; other cell types will become available in the near future. Allele Biotechnology has commenced differentiating iPSCs along several lineages using our own iPSCs of superior quality.
With cells of disease-matching tissue types derived from patients’ iPSCs, cell-based assays can be designed and developed using various assay formats. Allele Biotech’s leading capacities in fluorescence and bioluminescence, gene silencing, delivery vehicles and single-domain targeting agents will be of unmatched value to drug discovery partners.
1. Review: Wu, SM and Hochedlinger, K. “Harnessing the potential of induced pluripotent stem cells for regenerative medicine ” 2011, Nature Cell Biology, V13-5, 497-505.
2. Allele Biotech publication: Warren, L., Ni, Y., Wang, J. and Guo, X. “Feeder-Free Derivation of Human Induced Pluripotent Stem Cells with Messenger RNA” 2012, Nature’s Scientific Reports, doi:10.1038/srep00657.
For business development contact:
iPS@allelebiotech.com
858-587-6645
Fax 858-587-6692
www.allelebiotech.com
6404 Nancy Ridge Drive
San Diego, CA 92121
Related products for academic customers: Non-Integrating iPSC Generation Product Line http://www.allelebiotech.com/non-integrating-ipsc-generation/
New Product of the week: 6F mRNA Reprogramming Premix: $995 for 10 reprogramming!
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008