When Great is not Good Enough—VHH Antibodies Engineered for 10 Fold Affinity Increase
Single Domain antibodies (VHH fragments, nanobodies, or as we call them, nAbs) have been generated by injecting llamas with ligand-bound GPCR for the purpose of obtaining crystals of active-state structures. Such structural information could be critical in understanding drug functions and screening for new drugs. The unique ability of VHH fragments to fit into protein-protein complex crevices and hold proteins together was demonstrated by two Nature publications from Brian Kobilka’s group at Stanford ([1, 2], also see Allele Newsletter of Sep 4th, 2013). The nano antibody used in those studies, Nb80, showed affinity towards only the active state of the target GPCR.
However, even with an antibody as great as Nb80, the authors were only able to co-crystal GPCR beta2-adrenoceptor (b2AR) with high affinity agonists, not its natural agonists such as adrenaline. In yet another Nature paper published just now, the Kobilka lab showed that Nb80 could be further improved by 10 times in affinity, through in vitro evolution [3]. They presented Nb80 on the surface of yeast using an existing yeast display system, then applied standard limited mutagenesis and magnetic separation technologies for screening. After about 5 rounds of selection, a new version of VHH Nb6B9 was isolated that bound to ligand-loaded GPCR with a kD of 6.4 nM. For the first time, a co-crystal of b2AR-adrenoline was made.
Rasmussen et al. Nature, 2011 Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Rasmussen et al. Nature, 2011 Crystal structure of the b2 adrenergic receptor–Gs protein complex
Ring et al. Nature, 2013 Adrenaline-activated structure of b2-adrenoceptor stabilized by an engineered nanobody
Update here http://www.allelebiotech.com/nab
Video Blog: Beating Heart Cells from Stem Cells
Click on the video link, and watch carefully. You will see the small patches of the iPSC-derived cardiomyocytes beat, like tiny hearts.
Beating heart cells from stem cells
Is “100% Conversion” No Longer a Taboo Phrase in the iPSC Field?
“By removing a single protein called Mbd3, a team at the Weizmann Institute of Science in Rehovot, Israel, was able to increase the conversion rate to almost 100%”. A staggering statement written by Monya Baker for Nature News, after her journal’s online publication by Rais et al. reported their discovery. This discovery displays that by removing the expression of nucleosome remodeling and deacetylation repressor complex member Mbd3, all cells can be reprogrammed with the OSKM Yamanaka factors. What we have described last September in Nature’s Scientific Reports (Warren et al. 2012) that by fusing Oct4 with strong transcription activation domain, we achieved essentially the same results which we call “bulk conversion”. One can imagine that the strong transcription activation domain very likely effects such strong reprogramming through some type of chromosomal remodeling indirectly.
As we later learned, people in the field believe that reprogramming is a stochastic event and it is impossible to achieve something like “bulk conversion”. We are happy to see that sea change now that a separate group also demonstrated “bulk conversion”. Recently we have shown that instead of 2 weeks or more, as required when mRNA reprogramming was first reported or commonly practiced, we could start taking up iPSCs around a week without doing any traditional transfection—just change the medium everyday as for all stem cell cultures, with mRNA complex as a supplement to be added together with bFGF.
Knocking-down Mbd3 is potentially useful, especially in situations where we have difficulty reprogramming a particular line, and it could make reprogramming more successful in more demanding conditions such as leaving out B18R as we recently established. However one needs to be cautious about the unavoidable off-target effects of siRNAs when including siRNA against Mbd3 in their RNA mix.
Rais et al. Nature (2013)
mRNA Delivery And the Next Wave of Regenerative Medicine
Published online by Nature Biotechnology, researchers from Ken Chien’s lab at Harvard and other coauthors showed that modified mRNA of VEGF-A injected intramyocardially resulted in the expansion and directed differentiation of endogenous heart progenitors. VEGF-A modRNA markedly improved heart function and enhanced long-term survival of recipients by directing epicardial progenitor cells toward cardiovascular cell types. This publication appears to be the first example of using mRNA as a delivery platform for cell fate-related therapy. AstraZeneca recently invested $240 million on mRNA-related delivery via Moderna, a company with roots within the Harvard stem cell group.
The drastically increased efficacy of using the mRNA platform was accredited to the pulse-like kinetics of mRNA expression profile. It was explained by the fact that native paracrine signals are often transient and precisely regulated in time and space, therefore the pulse-like expression profile of modRNA might be well suited to delivering paracrine-factor signals. Transfected mRNA molecules do not need to penetrate the nuclear membrane, which greatly enhances the efficiency of protein expression on a per transfected molecule over DNA. mRNAs turn over in a much faster pace than plasmid-mediated transgene expression. This is beneficial to many cell fate decisions as exemplified by this recent publication.
Allele Biotech’s reprogramming technologies, licensed by some of the leading stem cell therapy companies, are built around the mRNA platform. We chose mRNA as our core technology to not only change cell fate, but also direct differentiation. We know this platform is the future for cell fate manipulation because we have seen how robustly mRNA expression made the day-and-night difference in gene expression when compared to plasmid DNA (episomal or not), retrovirus, lentivirus, baculo virus, or even transfected proteins. We could convert human fibroblasts into iPSCs, in bulk, in as short as one week with no more effort than changing mRNA complex-containing medium.
Another recent development in iPSC research is in situ reprogramming. Abad et al. generated mice carrying a Tet-inducible cassette of the four cell-reprogramming factors. They then added feed doxycycline to the animals. After several weeks, teratomas appeared in various tissues, indicating that in situ reprogramming had occurred. The iPSCs created this way did not appear to have much advantage over in vitro produced iPSCs other than they are totipotent (helpful if you are studying placenta). Nevertheless, the concept of changing cell fate in situ as dramatically as complete reprogramming is an important leap of faith. As for the next big step, it is easy to see that mRNAs are well suited for in situ reprogramming, as well as transdifferentiation, and more complex gene delivery than the above mentioned VEGF-A alone in heart treatment.
References:
Zangi et al. Nature Biotechnology, http://www.nature.com/nbt/journal/vaop/ncurrent/abs/nbt.2682.html
Abad et al. Nature, http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature12586.html
Picture Blog: How Do You Like Your iPSCs, Clonal or Bulk-Conversion?
Reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs) is commonly considered a stochastic process, i.e. with randomness, which offers an excuse for the commonly seen low efficiency and low constancy of making iPSCs. We have demonstrated time and again that by using potent mRNA cocktails, the majority of the fibroblasts seeded in a well can be converted into pluripotent stage in a nearly synchronized manner (Warren et al. 2012, Warren and Wang 2013, and this Allele Picture Blog series). mRNA molecules can function robustly yet transiently while avoiding the need of entering the nucleus, a bottle-neck for all DNA-based vehicles.
Other researchers are used to the idea of clonal expansion partly because isolating iPSCs from “clones” was a common step during reprogramming using viruses or other low efficiency methods, even though those clones were not necessarily from single precursor cells. This week, the Allele iPSC team developed a new way of managing our mRNA reprogramming that allowed us to achieve clonal iPSCs that appear to be a lot purer and more likely true clones compared to previous reports, without compromising any of the main benefits of our protocol, e.g. feeder-free, xeno-free, footprint-free, very fast and highly efficient. This work is currently supported by an NIDA/NIH grant to Dr. Jiwu Wang at Allele Biotech.
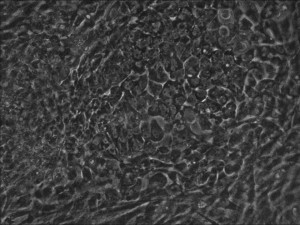
Traditional bulk-conversion by the Allele mRNA reprogramming protocol developed by Warren et al. The picture shows large patches of cells becoming stem cells almost overnight around the 9th day of adding mRNA-cocktail supplement to the media.
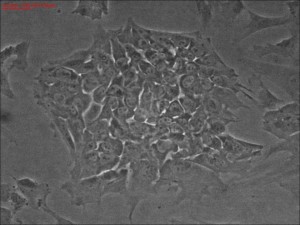
Clonal iPSC formation using a modified mRNA reprogramming protocol. The picture shows a typical clone of stem cells that originated from likely single cells.
Warren, Ni, Wang, and Guo, Scientific Reports, 2012
Warren and Wang, Current Protocols, 2013, in press
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008



