footprint free
cGMP Compliance: What Does It Mean for Your Cell Lines?
As the promise for cell-based therapy grows, the interest in making clinically relevant cell lines has skyrocketed for industrial and academic researchers alike. For translation into human therapies, cell-based products must be made following current Good Manufacturing Practice (cGMP). Many groups have already claimed to generate cell lines that are “cGMP-compliant,” “cGMP-ready,” or “certifiable under cGMP.” But what does it take to be truly cGMP-compliant, and what practices can you introduce in your lab to comply with cGMP standards?
A common misconception in the United States is that a facility is granted a ‘cGMP license’ from the government to manufacture cGMP-grade products. Rather, the Food and Drug Administration (FDA) evaluates the manufacturing process for each product to determine if it is compliant with cGMP standards. The primary concern when it comes to deriving cell-based products for therapies is making sure that the product is derived in a safe and reproducible manner. To ensure maximum quality assurance, researchers should
• choose reliable, xenogeneic-free raw materials,
• establish and monitor a clean environment,
• qualify all equipment and software,
• remove variation in laboratory procedures by creating detailed Standard Operating Procedures (SOPs) and by providing rigid process validation at each step.
Nevertheless, even establishing robust quality assurance does not imply that the process is scalable for commercial production. In the world of biologics, “the product is the process.” A requisite step to ensure a smooth transition to cGMP practice is to ensure that the process of manufacturing is not altered due to changes in production scale. For example, depending on the therapy, millions or billions of cells may be required for a single patient. Therefore, it is in the best interest of the researchers to develop a scalable method at the beginning to avoid revamping the entire process (e.g., changing from adherent cells to suspension). Along these lines, the quality control (QC) requirements of cell-based products should be carefully considered and not have to include difficult-to-assay tests. For example, some cell lines have been qualified as cGMP-compliant upon conversion from research-grade conditions to cGMP quality standards. Rigorous tests were performed on the converted lines to ensure that the cells were free of contamination. Even though strict measures were carried out to ensure cGMP compliancy, deriving cell lines in this manner makes scalability and reproducibility a challenge. Ideally, the entire process of deriving cell products for clinical use should be performed under cGMP conditions: from the acquisition of human tissue to the manufacturing, testing, and storage of derivative cell products.
Another important consideration when instituting cGMP-compliance is documentation. Each process must be described with rigorous SOPs, the training of individual manufacturing operators must be well-documented, and the entire established process must be validated and well noted. Failure to document—in the eyes of the FDA—is often equated with failure to perform the underlying activity. It is equally important to remain ‘current.’ The FDA expects manufacturing processes to stay up-to-date with current regulations, even as policies change.
For an academic lab, closely aligning with cGMP standards can ensure that the resulting cell lines are comparable to other truly cGMP-produced products used during clinical trials. It is in the best interest of academic researchers to establish rigorous SOPs and use qualified reagents and equipment, even if it is not possible to carry out all steps in a certified cleanroom. Whenever possible, it is advisable to acquire truly cGMP cell lines from appropriate sources for preclinical projects; if prohibited by costs or other reasons, it is recommended to use a protocol that is as close to cGMP as possible.
New Allele Biotech Publication on Stem Cells
Feeder-Free Reprogramming of Human Fibroblasts with Messenger RNA
Current Protocols in Stem Cell Biology • November 13, 2013
DOI: 10.1002/9780470151808.sc04a06s27
Authors: Luigi Warren, Jiwu Wang
This unit describes a feeder-free protocol for deriving induced pluripotent stem cells (iPSCs) from human fibroblasts by transfection of synthetic mRNA. The reprogramming of somatic cells requires transient expression of a set of transcription factors that collectively activate an endogenous gene regulatory network specifying the pluripotent phenotype. The necessary ectopic factor expression was first effected using retroviruses; however, as viral integration into the genome is problematic for cell therapy applications, the use of footprint-free vectors such as mRNA is increasingly preferred. Strong points of the mRNA approach include high efficiency, rapid kinetics, and obviation of a clean-up phase to purge the vector. Still, the method is relatively laborious and has, up to now, involved the use of feeder cells, which brings drawbacks including poor applicability to clinically oriented iPSC derivation. Using the methods described here, mRNA reprogramming can be performed without feeders at much-reduced labor and material costs relative to established protocols.
Allele iPSC Service and Technology Licensing Contact: http://www.allelebiotech.com/cell-line-and-culture-services/#ips-line
New Allele Product of the Month: FP-nAb™ products for 100% pull-down
Picture Blog: How Do You Like Your iPSCs, Clonal or Bulk-Conversion?
Reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs) is commonly considered a stochastic process, i.e. with randomness, which offers an excuse for the commonly seen low efficiency and low constancy of making iPSCs. We have demonstrated time and again that by using potent mRNA cocktails, the majority of the fibroblasts seeded in a well can be converted into pluripotent stage in a nearly synchronized manner (Warren et al. 2012, Warren and Wang 2013, and this Allele Picture Blog series). mRNA molecules can function robustly yet transiently while avoiding the need of entering the nucleus, a bottle-neck for all DNA-based vehicles.
Other researchers are used to the idea of clonal expansion partly because isolating iPSCs from “clones” was a common step during reprogramming using viruses or other low efficiency methods, even though those clones were not necessarily from single precursor cells. This week, the Allele iPSC team developed a new way of managing our mRNA reprogramming that allowed us to achieve clonal iPSCs that appear to be a lot purer and more likely true clones compared to previous reports, without compromising any of the main benefits of our protocol, e.g. feeder-free, xeno-free, footprint-free, very fast and highly efficient. This work is currently supported by an NIDA/NIH grant to Dr. Jiwu Wang at Allele Biotech.
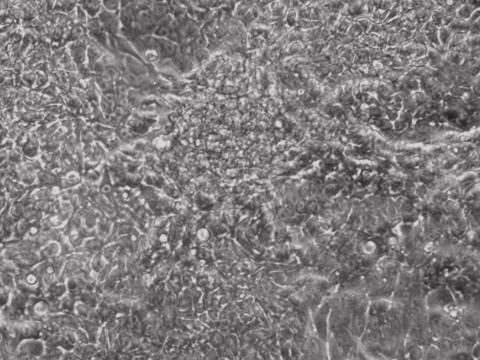
Traditional bulk-conversion by the Allele mRNA reprogramming protocol developed by Warren et al. The picture shows large patches of cells becoming stem cells almost overnight around the 9th day of adding mRNA-cocktail supplement to the media.
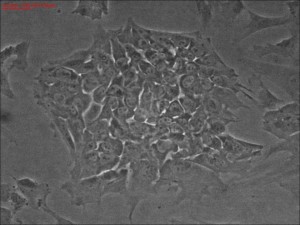
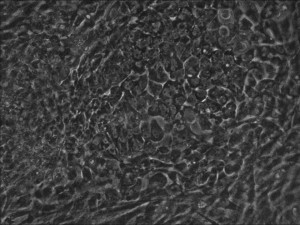
Clonal iPSC formation using a modified mRNA reprogramming protocol. The picture shows a typical clone of stem cells that originated from likely single cells.
Warren, Ni, Wang, and Guo, Scientific Reports, 2012
Warren and Wang, Current Protocols, 2013, in press
Picture Blog: More Efficient Reprogramming for Creating Induced Stem Cells (iPSCs)
Researchers at Allele Biotech achieved reprogramming of human fibroblasts into iPSCs within one week by including mRNA transfection supplement in daily medium change, at “bulk conversion” efficiency, and with cells seeded at a much wider density range compared to our previous publications. These significant improvements will further facilitate high throughput, large scale iPSC production using Allele’s feeder-free, xeno-free, footprint-free reprogramming, which was already a preferred method for both clinical applications and stem cell banking. The reprogramming project is currently being funded by the NIDA/NIH.
Allele Biotech Receives $200,000 Grant to Update Its mRNA Reprogramming Commercial Products and Services
On June 10, 2013 Allele received an SBIR award from the National Institute of Drug Abuse (NIDA/NIH) entitled “Revolutionary Technology for Efficient Derivation of Human iPSCs with Messenger RNA”. The goal of the proposed project is to provide to the biomedical research market an advanced reagent kit and services for highly efficient reprogramming of high quality human induced pluripotent stem cells (iPSCs). At the core of this kit is the Allele team’s recent development transcribed messenger RNA (mRNA). Compared to other reprogramming methods, such as lentivirus, Sendai virus, protein, small molecules or any combinations of these reagents, our new generation of the mRNA method often requires less than half the time while sometimes achieving “bulk conversion” efficiency.
While the Allele reprogramming technology was designed for clinical use as the process is feeder-free, xeno-free, chromosome integration-free, as well as without the need for cell splitting, PI, Dr. Jiwu Wang states, “Our purpose of executing the NIH-funded research it to make our method so easy that any researcher can integrate iPSC into his or her projects.” In addition to the extremely high efficiency, mRNA-generated iPSCs should also be more stable because there are no genetic alterations, more uniform among all clones as there is no clonal event, and ultimately suitable for future autologous cell therapy now that creating iPSCs from patient tissue cells should no longer be the rate-limiting steps.
Allele’s business model is to provide cGMP-grade iPSCs to pharmaceutical companies and perform large scale reprogramming by partnering first with university-affiliated hospitals. Great progress has been made in both directions, which has prompted the initiation of a cGMP unit within Allele’s newly acquired building in San Diego.
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008