neuron
Picture Blog: A Short Path from Human mRNA-iPSCs to Neurons in Record Speed
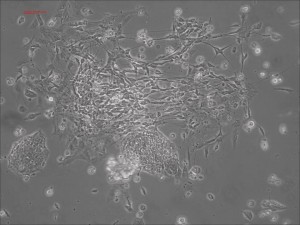
Traditional differentiation protocols use embryoid body (EB) formation as the first step of lineage restriction to mimic early human embryogenesis, which is then followed by manual selection of neuroepithelial precursors. This procedure is tedious and often inconsistent. We have developed a novel neural differentiation scheme that directs human iPSCs (created with the Allele 6F mRNA reprogramming kit) that progressed, as attached culture, to neural precursor cells (NPCs) in just 4-6 days, half the time it typically takes by other methods. From NPCs it takes about another 5-6 days for neural rosettes to form (see figures below); upon passage, cells in neural rosettes differentiate into neurons in 24 hours.
The neural progenitors at the rosettes stage can be stocked and expanded, before differentiated into different types of neurons. We are working on specifically and efficiently different these neural progenitor cells into dopaminergic, glutamatergic, GABAergic, and other types of sub-types of neurons with Allele’s technologies (Questions? email the Allele Stem Cell Group at iPSatAllelebiotech.com).
Neural rosettes formed efficiently in wells without going through EB.
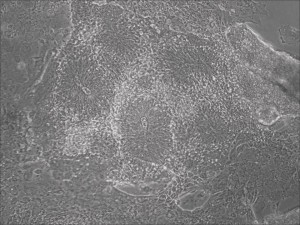
Human iPSC-derived neurons are created in a short regimen developed at Allele Biotech
Visualizing Endogenous Synaptic Proteins in Living Neurons
The recently published method is based on the generation of disulfide-free “intrabodies”, a structure from the 10th fibronectin type III domain known as FingRs. These affinity molecules were fused to GFP for direct fluorescence miscroscopy. The FingRs do not need di-sulfite bonds and are therefore better folders in mammalian cells. Specifically, a library was screened with in vitro display to identify FingRs that bind two synaptic proteins, Gephyrin and PSD95. After the initial selection, the researchers from USC secondarily screened binders using a cellular localization assay to identify potential FingRs that bind at high affinity in an intracellular environment. As it turned out, only 10-20% of the original positive clones bind well inside the cells, suggesting this type of further screening was a critical step.
The expression of intrabody is transcriptionally regulated by the target protein through a ZFN-repressor fusion. This transcriptional control system matches the expression of the intrabody to that of the target protein regardless of the target’s expression level. This design virtually eliminates unbound FingR, resulting in very low background that allows unobstructed visualization of the target proteins. As result, the FingRs presented in this study enabled live cell visualization of excitatory and inhibitory synapses, and apparently without affecting neuronal function.
Technically, the reason to use in vitro mRNA display was required by the need to use a large library (>10exp12, beyond the limit of the more commonly used phase display) to find good binders. A similar visualization system can be established using more potent affinity domains such as the VHH single-domain antibodies that have only one, sometimes dispensable, di-sulfite bond. The VHH domain nanobodies can be more easily isolated from camelid animals. Another improvement to the visualization system can be made by using stronger, superresolution-ready FPs such as mNeonGreen or mMaple to enable single molecule imaging, which is particularly interesting for studying synapses and applied to the BRAIN initiative.
Gross et al. Neuron, June 2013, http://www.ncbi.nlm.nih.gov/pubmed/23791193
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008



