mRNA
Picture Blog: A Short Path from Human mRNA-iPSCs to Neurons in Record Speed
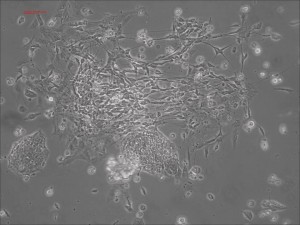
Traditional differentiation protocols use embryoid body (EB) formation as the first step of lineage restriction to mimic early human embryogenesis, which is then followed by manual selection of neuroepithelial precursors. This procedure is tedious and often inconsistent. We have developed a novel neural differentiation scheme that directs human iPSCs (created with the Allele 6F mRNA reprogramming kit) that progressed, as attached culture, to neural precursor cells (NPCs) in just 4-6 days, half the time it typically takes by other methods. From NPCs it takes about another 5-6 days for neural rosettes to form (see figures below); upon passage, cells in neural rosettes differentiate into neurons in 24 hours.
The neural progenitors at the rosettes stage can be stocked and expanded, before differentiated into different types of neurons. We are working on specifically and efficiently different these neural progenitor cells into dopaminergic, glutamatergic, GABAergic, and other types of sub-types of neurons with Allele’s technologies (Questions? email the Allele Stem Cell Group at iPSatAllelebiotech.com).
Neural rosettes formed efficiently in wells without going through EB.
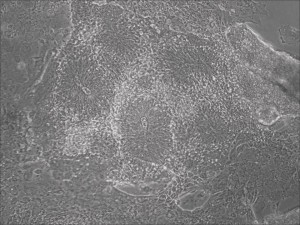
Human iPSC-derived neurons are created in a short regimen developed at Allele Biotech
mRNA Delivery And the Next Wave of Regenerative Medicine
Published online by Nature Biotechnology, researchers from Ken Chien’s lab at Harvard and other coauthors showed that modified mRNA of VEGF-A injected intramyocardially resulted in the expansion and directed differentiation of endogenous heart progenitors. VEGF-A modRNA markedly improved heart function and enhanced long-term survival of recipients by directing epicardial progenitor cells toward cardiovascular cell types. This publication appears to be the first example of using mRNA as a delivery platform for cell fate-related therapy. AstraZeneca recently invested $240 million on mRNA-related delivery via Moderna, a company with roots within the Harvard stem cell group.
The drastically increased efficacy of using the mRNA platform was accredited to the pulse-like kinetics of mRNA expression profile. It was explained by the fact that native paracrine signals are often transient and precisely regulated in time and space, therefore the pulse-like expression profile of modRNA might be well suited to delivering paracrine-factor signals. Transfected mRNA molecules do not need to penetrate the nuclear membrane, which greatly enhances the efficiency of protein expression on a per transfected molecule over DNA. mRNAs turn over in a much faster pace than plasmid-mediated transgene expression. This is beneficial to many cell fate decisions as exemplified by this recent publication.
Allele Biotech’s reprogramming technologies, licensed by some of the leading stem cell therapy companies, are built around the mRNA platform. We chose mRNA as our core technology to not only change cell fate, but also direct differentiation. We know this platform is the future for cell fate manipulation because we have seen how robustly mRNA expression made the day-and-night difference in gene expression when compared to plasmid DNA (episomal or not), retrovirus, lentivirus, baculo virus, or even transfected proteins. We could convert human fibroblasts into iPSCs, in bulk, in as short as one week with no more effort than changing mRNA complex-containing medium.
Another recent development in iPSC research is in situ reprogramming. Abad et al. generated mice carrying a Tet-inducible cassette of the four cell-reprogramming factors. They then added feed doxycycline to the animals. After several weeks, teratomas appeared in various tissues, indicating that in situ reprogramming had occurred. The iPSCs created this way did not appear to have much advantage over in vitro produced iPSCs other than they are totipotent (helpful if you are studying placenta). Nevertheless, the concept of changing cell fate in situ as dramatically as complete reprogramming is an important leap of faith. As for the next big step, it is easy to see that mRNAs are well suited for in situ reprogramming, as well as transdifferentiation, and more complex gene delivery than the above mentioned VEGF-A alone in heart treatment.
References:
Zangi et al. Nature Biotechnology, http://www.nature.com/nbt/journal/vaop/ncurrent/abs/nbt.2682.html
Abad et al. Nature, http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature12586.html
Picture Blog — Making mRNAs by In Vitro Transcription for Transgene Expression and R-iPSCs
R-iPS Cell FAQ 2:
What is the expected yield from the in vitro trancription (IVT) reactions?
Performed as described, you should recover around 40 ug RNA from each 40 uL IVT reaction.
R-iPS Cell FAQ 3:
How can the success of the RNA synthesis protocol be assessed?
Run 500 ng (5 uL) of the concentration-adjusted products on an E-gel to check for consistent product yield and relative product sizes, and to confirm the absence of secondary bands or smears.
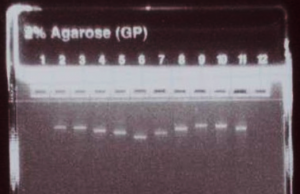
Making Transfection-Grade mRNA by IVT (In Vitro Transcription)
RNases are an often feared in molecular biology labs because of their high stability and ominous presence in virtually all living systems. Consequently, people who work with RNA are trained to exercise extreme caution to avoid RNA degradation: change gloves often because human hands ooze RNases; use only sterilized labware as microbes may be sources of RNases; for surfaces that can’t be autoclaved, use sprays like “RNase Zap” (SDS- or guanidine-containing solutions). Such cautionary steps are especially necessary when dealing with low abundance RNA samples.
RNAs can be produced by in vitro transcription (IVT), a simple reaction requiring only a DNA template (double-stranded or even single-stranded DNA as long as the promoter region is double-stranded), RNA polymerase (from T7, SP6, or T3 phage), NTPs, and a reaction buffer that provides appropriate salt and pH. Standard NTPs may be replaced with modified ones to either increase stability or to reduce immune-response when transfected into cultured cells. Additionally, a 5’ cap structure may be added during IVT for further stabilizing mRNAs inside the cells post transfection. Using a commercially assembled kit, one can routinely produce 40-50 µg of mRNA from 1 µg of DNA template in a single 20-50 µl reaction.
At such high concentrations, IVT mRNAs are not nearly as sensitive to RNase-mediated degradation as low-abundance samples. The mRNA can be easily observed on agarose gels that are regularly used for DNA, and their integrity can be monitored after transcription or storage. In most cases one distinct band of mRNA from an IVT reaction is obtained as long as a clean DNA template is used. Preparing a good, uniform IVT template is critical to prevent aberrant products. By using high quality templates, IVT mRNA produced in your own lab are often higher in quality than mRNAs purchased from current commercial sources (Figure in Blog shows mRNAs generated by IVT for R-iPSC). Sometimes there are minor bands created during IVT, but they normally do not interfere with the intended uses of the mRNA, and can be purified away with a purification kit (by using a discriminating purification scheme such as Allele Biotech’s Surface Bind RNA Purification, smaller species can be specifically removed, a separate topic for another blog).
Once produced, mRNAs can be stored at -20C for months, or -80C nearly indefinitely.
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008




