integration-free
Picture Blog: How Do You Like Your iPSCs, Clonal or Bulk-Conversion?
Reprogramming of differentiated cells into induced pluripotent stem cells (iPSCs) is commonly considered a stochastic process, i.e. with randomness, which offers an excuse for the commonly seen low efficiency and low constancy of making iPSCs. We have demonstrated time and again that by using potent mRNA cocktails, the majority of the fibroblasts seeded in a well can be converted into pluripotent stage in a nearly synchronized manner (Warren et al. 2012, Warren and Wang 2013, and this Allele Picture Blog series). mRNA molecules can function robustly yet transiently while avoiding the need of entering the nucleus, a bottle-neck for all DNA-based vehicles.
Other researchers are used to the idea of clonal expansion partly because isolating iPSCs from “clones” was a common step during reprogramming using viruses or other low efficiency methods, even though those clones were not necessarily from single precursor cells. This week, the Allele iPSC team developed a new way of managing our mRNA reprogramming that allowed us to achieve clonal iPSCs that appear to be a lot purer and more likely true clones compared to previous reports, without compromising any of the main benefits of our protocol, e.g. feeder-free, xeno-free, footprint-free, very fast and highly efficient. This work is currently supported by an NIDA/NIH grant to Dr. Jiwu Wang at Allele Biotech.
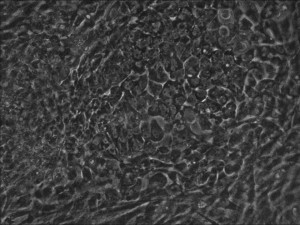
Traditional bulk-conversion by the Allele mRNA reprogramming protocol developed by Warren et al. The picture shows large patches of cells becoming stem cells almost overnight around the 9th day of adding mRNA-cocktail supplement to the media.
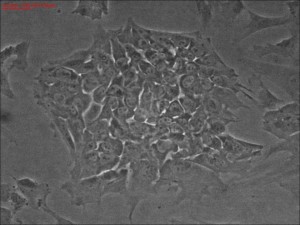
Clonal iPSC formation using a modified mRNA reprogramming protocol. The picture shows a typical clone of stem cells that originated from likely single cells.
Warren, Ni, Wang, and Guo, Scientific Reports, 2012
Warren and Wang, Current Protocols, 2013, in press
Expression of iPS Factors from Transfected mRNA
Differentiated cells can be reprogrammed to pluripotency by enforced expression of certain combinations of stem cell-specific protein factors in them. The power of this method was first demonstrated by Yamanaka’s group using retroviruses carrying Oct3/4, Sox2, c-Myc, and Klf4. Alternative factors such as Lin28 and Nanog, and additional factors such as the human telomerase gene hTert and shRNA against p53 were also shown to contribute to reprogramming. From the very beginning it was realized that viral integration would pose a major problem in using the induced pluripotent stem cells (iPSCs) for clinical purposes. There have been multiple attempts to circumvent this problem by using non-integrating vectors such as plasmid, minicircle DNA, adenovirus, baculovirus, removable transposons, episomal DNA, or by introducing recombinant proteins with a transmembrane domain into target cells. From reports in the field and customer feedbacks it seems that retroviral or lentiviral systems are still the most efficient in reprogramming. mRNA is about the only option left unreported, until an article by Warren et al was published in Cell Stem Cell online recently.
From that report, it is clear that the reason that it took so long for RNA-induced iPSCs (RiPSCs) to appear in the literature was because synthetic mRNAs activate interferon responses in mammalian cells, reminding us of the early days of RNAi. The authors took a number of steps to reduce interferon responses, including adding a 5’-cap (actually a fairly standard step in in vitro transcription), using a phosphatase to remove 5’ triphosphates on uncapped mRNAs, and using modified C and U bases (5-methucytidine or 5mC and pseudouridine or psi) during T7 promoter-driven in vitro transcription. The prepared mRNA was then administered everyday for 17 days at an amount not clearly defined in the paper. The main benefit of this method is of course that there is no gene integration to alter the chromosome. The efficiency of the new method was also compared to using viral vectors and it was shown that 1.4% conversion efficiency was achieved vs retroviral systems’ 0.01% (although we have experienced better results using lentivirus, at least the 4-in-1 version).
The DNA templates used for in vitro transcription of the iPS factors were created by multiple PCR reactions and bridged ligation; it could also be done by other cloning strategies. For those excited about trying this new way of making iPSCs, the major hassle would be preparing modified mRNAs good and abundant enough for 17 consecutive transfections. Allele Biotech would like to provide custom services, before offering shelf products, for creating such mRNAs as the method sounds potentially very helpful to many researchers in the iPSC field.
- New Product of the Week 100410-101010:
pLICO-mWasabi (Promoterless FP Reporter Vector ), listed as product-on-demand, now available, ABP-HL-PE40010 $395.00.
- Promotion of the Week 100410-101010:
Barrier too high to start using virus? Allele lowers it for starters, $500 for bactulo virus protein production, and $300 retrovirus packaging. Code 100310VIVEC, email vivec@allelebiotech.com
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008



