footprint free
Conducting Massively Parallel Sequencing
One of the major breakthroughs in modern biology is the development of massively parallel sequencing, also called next generation sequencing (NGS), which enabled the complete delineation of the human genome more than a decade ago. Since then many more species’ genomes have been sequenced, and the cost per genome has dropped from billions to mere thousands of dollars. New discoveries are being made as a result of the capability many research teams now possess to not only sequence chromosomal DNA, but also to identify which regions a protein of interest specifically binds (Chip-seq), analyze a whole transcriptome of a cell population under investigation (RNA-seq), or find out which RNA regions an RNA binding protein resides (CLIC-seq).
While it is inevitable that many PIs will seriously consider the inclusion of deep sequencing in their next grant proposal, it is not necessarily easy to take the first step and get their feet wet, so to speak. Knowing what format (e.g. 454 for longer reads, HighSeq for higher accuracy, or Ion Torren for bench top convenience) to use and how much to pay requires a vast amount of knowledge and experience. Even when you are done with sample prep, amplification and sequencing, to handle such massive amount of data is not trivial—transporting data alone can be a headache. A database server for storage and analysis requires another layer of expertise. There is no easy solution but to get started somehow. However, be prepared to deal with these issues.
Whether the cost on a type of next generation service is justifiable depends on whether it is required for your purposes. For example, when analyzing a person’s propensity of developing a disease by using known, disease-relevant genetic information, often times exome sequencing is sufficient. This costs anywhere between $1,000 to $3,000 with 100X coverage, significantly less than sequencing a complete genome which typically costs ~$5,000 at ~20x coverage.
High coverage sequencing of maternal blood DNA has been developed into clinically approved prenatal diagnosis of trisomy in Down’s syndrome and other chromosomal abnormalities. Transcriptome analysis helped the understanding of how reprogramming works when iPSCs are. Looking forward, with more routine use of deep sequencing we can predict with much more certainty the “off-target” effects of RNAi or cellular toxicity of chromosomal modifications enabled by ZFN, TALEN, or CRISPR. As a matter of fact, we believe that transcriptome sequencing should be required after each RNAi event to prove a specific linkage between knockdown and functions; similarly, whole genome sequencing results need to be provided after making a site directed chromosomal change in the future for high level publications.
*This blog partially resulted from discussions between Jiwu Wang and his colleagues, who are NGS experts at UCSD’s Cellular and Molecular Medicine, Moore Cancer Center, and BGI Americas.
Picture Blog — Making mRNAs by In Vitro Transcription for Transgene Expression and R-iPSCs
R-iPS Cell FAQ 2:
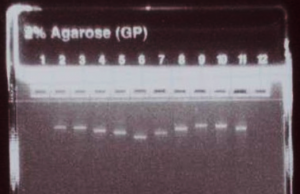
What is the expected yield from the in vitro trancription (IVT) reactions?
Performed as described, you should recover around 40 ug RNA from each 40 uL IVT reaction.
R-iPS Cell FAQ 3:
How can the success of the RNA synthesis protocol be assessed?
Run 500 ng (5 uL) of the concentration-adjusted products on an E-gel to check for consistent product yield and relative product sizes, and to confirm the absence of secondary bands or smears.
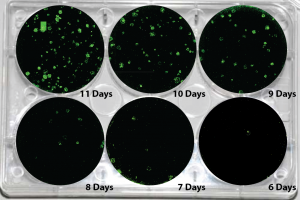
Picture Blog — Human mRNA-Induced Pluripotent Stem Cells Generated in Days
R-iPS Cell FAQ 1:
What phenotypic changes can be observed during a successful reprogramming trial?
About one week out, target fibroblasts should show an involution of fibroblastic processes, and foci or clusters of epitheliod cells—ideally with small nuclei, minimal cytoplasm, and signs of ongoing mitosis—should appear. Colonies with hESC morphology typically start emerging in ~10-14 days.
Fusion of the Transcription Domain to iPS Factors Radically Enhances Reprogramming
Induced pluripotent stem cells (iPSCs) can be achieved through introduction of a small group of stem cell specific transcription factors. Ever since this was first demonstrated by Takahashi and Yamanaka, there have been relentless efforts for improving the efficiency of this generally inefficient process. There is also a general opinion that iPSCs are different from each other and from embryonic stem cells (ESCs) in various aspects, depending on the method of the induction. As a result, another focus of the reprogramming field has been to find ways for creating iPSCs that are as close to ESCs as possible. One of the parameters for defining stem cell status is their epigenetic characters; epigenetic changes have been demonstrated to occur during reprogramming of subsequent differentiation.
In fact, it seems that reprogramming can be largely described as a process composed of chromatin remodeling and specific transcription activation. Strong transcription activators are known to effectively recruit multiple chromatin remodeling complexes when exerting their functions. A good example is MyoD, a master transcription factor for skeletal myogenesis that can “single-handedly” switch (transdifferentiate) the fate of differentiated cells. Hirai et al. speculated that since MyoD is such a strong transcription factor, it may be able to increase chromatin accessibility to iPS factors if fused together. When transduced on retroviral vectors, Oct-TAD (Transcription Activation Domain) of MyoD, in combination with Sox2 and Klf4, increased the number of iPSC colonies by 40-fold. Additionally, these iPSCs appeared to quickly adopt stem cell gene expression profiles, days faster than when traditional Oct4, Sox2, c-Myc, and Klf4 were used; and sometimes the levels of pluripotency genes even exceeds those seen in ESCs. Amazingly, when using the fusion assisted method some colonies are formed without the help of feeder cells, a requirement of ESCs grown in similar medium. Does this mean that these iPSCs can even be more “stem-like” than embryonic stem cells?
Like MyoD, VP16, also widely known for its strong transcription activation domain, when fused to iPS factors, was shown to exhibit a similar stimulation effect on reprogramming. Although the details of the fusion arrangements and specificity appear to differ between MyoD and VP16, the fact that two research groups could achieve similar results using comparable strategies provides a good argument that other labs should at least consider this method when creating mouse or human iPSCs. Previously in our blog we have discussed using iPS factor mRNAs, a method originally developed by Warren et al., for substantially shortening the time required for reprogramming and making it more robust across cell types and media conditions. If the new TAD-fusion factors are used also in the mRNA format, then the protocol might be further shortened and simplified. If successful, this non-integrating approach could become a dominant method in the field, even making competitive non-integrating method such as Sendai and plasmid-based miRNA irrelevant.
New product of the week:SurfaceBind RNA purification system, higher capacity and simpler procedure than Qiagen or Ambion’s comparable products, particularly suitable for mRNA cleanup after in vitro transcription.
Promotion of the week: 30% off RNA SurfaceBind Purification kits. To redeem this offer email the code PURIFY to oligo@allelebiotech.com.
Categories
- Allele Mail Bag
- cGMP
- Customer Feedback
- Fluorescent proteins
- iPSCs and other stem cells
- nAb: Camelid Antibodies, Nanobodies, VHH
- Next Generation Sequencing (NextGen Seq)
- NIH Budget and You
- oligos and cloning
- Open Forum
- RNAi patent landscape
- SBIR and Business issues
- State of Research
- Synthetic biology
- Uncategorized
- Viruses and cells
- You have the power
Archives
- October 2018
- April 2018
- March 2018
- January 2018
- October 2017
- September 2017
- August 2017
- March 2017
- February 2017
- January 2017
- November 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- February 2016
- October 2015
- September 2015
- August 2015
- June 2015
- March 2015
- January 2015
- December 2014
- March 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- May 2012
- April 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- January 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- October 2008
- August 2008
- July 2008